Python Worksheet¶
Setup¶
Import packages¶
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
import statsmodels.api as sm
import statsmodels.formula.api as smf
# Remove unrelated warning
import warnings
warnings.simplefilter(action="ignore", category=FutureWarning)
/usr/local/lib/python3.7/dist-packages/statsmodels/tools/_testing.py:19: FutureWarning: pandas.util.testing is deprecated. Use the functions in the public API at pandas.testing instead.
import pandas.util.testing as tm
Download OASIS Dataset¶
We’ll be downloading the data from http://www.oasis-brains.org/pdf/oasis_longitudinal.csv.
A full explanation of the data can be found in:
Open Access Series of Imaging Studies (OASIS): Longitudinal MRI Data in Nondemented and Demented Older Adults. Marcus, DS, Fotenos, AF, Csernansky, JG, Morris, JC, Buckner, RL, 2010. Journal of Cognitive Neuroscience, 22, 2677-2684. doi: 10.1162/jocn.2009.21407
df = pd.read_csv('http://www.oasis-brains.org/pdf/oasis_longitudinal.csv')
# Print out the top 5 rows of the DataFrame
df.head(5)
| Subject ID | MRI ID | Group | Visit | MR Delay | M/F | Hand | Age | EDUC | SES | MMSE | CDR | eTIV | nWBV | ASF | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | OAS2_0001 | OAS2_0001_MR1 | Nondemented | 1 | 0 | M | R | 87 | 14 | 2.0 | 27.0 | 0.0 | 1987 | 0.696 | 0.883 |
| 1 | OAS2_0001 | OAS2_0001_MR2 | Nondemented | 2 | 457 | M | R | 88 | 14 | 2.0 | 30.0 | 0.0 | 2004 | 0.681 | 0.876 |
| 2 | OAS2_0002 | OAS2_0002_MR1 | Demented | 1 | 0 | M | R | 75 | 12 | NaN | 23.0 | 0.5 | 1678 | 0.736 | 1.046 |
| 3 | OAS2_0002 | OAS2_0002_MR2 | Demented | 2 | 560 | M | R | 76 | 12 | NaN | 28.0 | 0.5 | 1738 | 0.713 | 1.010 |
| 4 | OAS2_0002 | OAS2_0002_MR3 | Demented | 3 | 1895 | M | R | 80 | 12 | NaN | 22.0 | 0.5 | 1698 | 0.701 | 1.034 |
Pre-processing the Data¶
Dementia Status from CDR¶
We’re told in the associated paper (linked above) that the CDR values correspond to the following levels of severity:
0 = No dementia
0.5 = Very mild Dementia
1 = Mild Dementia
2 = Moderate Dementia
3 = Severe Dementia
So let’s derive this “Status” from the CDR values:
# Create a mapper from values to status/severity
cdr_mapper = {
0: "No Dementia",
0.5: "Very Mild Dementia",
1: "Mild Dementia",
2: "Moderate Dementia",
3: "Severe Dementia",
}
# Create a column, mapping CDR values to this status
# Convert to categorical type, with severity as order
df["Status"] = pd.Categorical(
df["CDR"].map(cdr_mapper),
categories=cdr_mapper.values(),
ordered=True
)
# Create colours for the different 'Status' categories
status_colours = {
i:j for i,j in zip(cdr_mapper.values(), sns.color_palette("colorblind", 5))
}
Now we can look at this status, and how many records there are both overall, and at each of the visits (or time points).
# Let's look at how many records we have of each severity
df["Status"].value_counts()
No Dementia 206
Very Mild Dementia 123
Mild Dementia 41
Moderate Dementia 3
Severe Dementia 0
Name: Status, dtype: int64
# There are no records for "Severe Dementia", so we can tidy things up by removing this unused category
df["Status"] = df["Status"].cat.remove_unused_categories()
# How many of each status do we have at each visit?
df.groupby(["Visit", "Status"]).size()
Visit Status
1 No Dementia 85
Very Mild Dementia 52
Mild Dementia 13
Moderate Dementia 0
2 No Dementia 73
Very Mild Dementia 49
Mild Dementia 19
Moderate Dementia 3
3 No Dementia 34
Very Mild Dementia 18
Mild Dementia 6
Moderate Dementia 0
4 No Dementia 10
Very Mild Dementia 3
Mild Dementia 2
Moderate Dementia 0
5 No Dementia 4
Very Mild Dementia 1
Mild Dementia 1
Moderate Dementia 0
dtype: int64
Variable Types¶
df.dtypes
Subject ID object
MRI ID object
Group object
Visit int64
MR Delay int64
M/F object
Hand object
Age int64
EDUC int64
SES float64
MMSE float64
CDR float64
eTIV int64
nWBV float64
ASF float64
Status category
dtype: object
Note: Further coercing of types (e.g. “Visit” is ordinal) is good practice, but the necessity depends on your analysis.
Convert Categorical Data¶
df["Sex"] = df["M/F"].map({
"M": 0,
"F": 1
})
Visits & Baseline Data¶
df["Visit"].value_counts()
1 150
2 144
3 58
4 15
5 6
Name: Visit, dtype: int64
Filter for baseline data, and look at their status on arrival
df_baseline = df.loc[df["Visit"]==1]
df_baseline["Status"].value_counts()
No Dementia 85
Very Mild Dementia 52
Mild Dementia 13
Moderate Dementia 0
Name: Status, dtype: int64
Data Exploration & Analysis¶
Summarize Data¶
df.describe(include="all")
| Subject ID | MRI ID | Group | Visit | MR Delay | M/F | Hand | Age | EDUC | SES | MMSE | CDR | eTIV | nWBV | ASF | Status | Sex | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| count | 373 | 373 | 373 | 373.000000 | 373.000000 | 373 | 373 | 373.000000 | 373.000000 | 354.000000 | 371.000000 | 373.000000 | 373.000000 | 373.000000 | 373.000000 | 373 | 373.000000 |
| unique | 150 | 373 | 3 | NaN | NaN | 2 | 1 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | 4 | NaN |
| top | OAS2_0048 | OAS2_0179_MR2 | Nondemented | NaN | NaN | F | R | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | No Dementia | NaN |
| freq | 5 | 1 | 190 | NaN | NaN | 213 | 373 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | 206 | NaN |
| mean | NaN | NaN | NaN | 1.882038 | 595.104558 | NaN | NaN | 77.013405 | 14.597855 | 2.460452 | 27.342318 | 0.290885 | 1488.128686 | 0.729568 | 1.195461 | NaN | 0.571046 |
| std | NaN | NaN | NaN | 0.922843 | 635.485118 | NaN | NaN | 7.640957 | 2.876339 | 1.134005 | 3.683244 | 0.374557 | 176.139286 | 0.037135 | 0.138092 | NaN | 0.495592 |
| min | NaN | NaN | NaN | 1.000000 | 0.000000 | NaN | NaN | 60.000000 | 6.000000 | 1.000000 | 4.000000 | 0.000000 | 1106.000000 | 0.644000 | 0.876000 | NaN | 0.000000 |
| 25% | NaN | NaN | NaN | 1.000000 | 0.000000 | NaN | NaN | 71.000000 | 12.000000 | 2.000000 | 27.000000 | 0.000000 | 1357.000000 | 0.700000 | 1.099000 | NaN | 0.000000 |
| 50% | NaN | NaN | NaN | 2.000000 | 552.000000 | NaN | NaN | 77.000000 | 15.000000 | 2.000000 | 29.000000 | 0.000000 | 1470.000000 | 0.729000 | 1.194000 | NaN | 1.000000 |
| 75% | NaN | NaN | NaN | 2.000000 | 873.000000 | NaN | NaN | 82.000000 | 16.000000 | 3.000000 | 30.000000 | 0.500000 | 1597.000000 | 0.756000 | 1.293000 | NaN | 1.000000 |
| max | NaN | NaN | NaN | 5.000000 | 2639.000000 | NaN | NaN | 98.000000 | 23.000000 | 5.000000 | 30.000000 | 2.000000 | 2004.000000 | 0.837000 | 1.587000 | NaN | 1.000000 |
Missing Data¶
# Count amount of missing data
df.isna().sum(0)
Subject ID 0
MRI ID 0
Group 0
Visit 0
MR Delay 0
M/F 0
Hand 0
Age 0
EDUC 0
SES 19
MMSE 2
CDR 0
eTIV 0
nWBV 0
ASF 0
Status 0
Sex 0
dtype: int64
Visualize Data¶
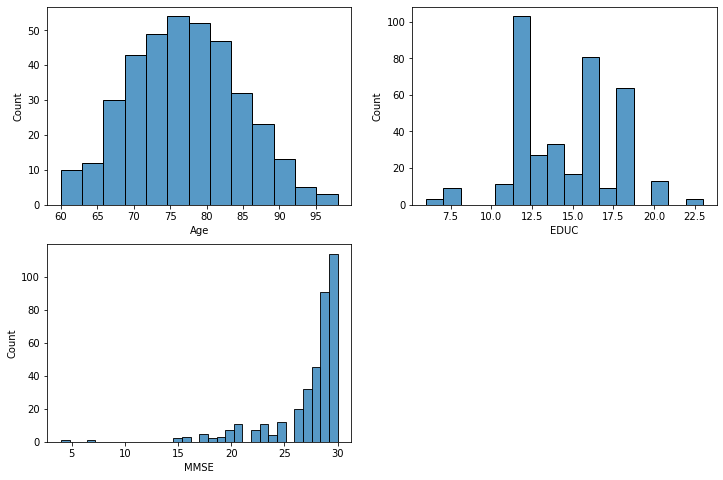
# Histograms
fig, axes = plt.subplots(2, 2, figsize=(12,8))
axes = axes.flatten()
for i, x_col in enumerate(["Age", "EDUC", "MMSE"]):
sns.histplot(data=df, x=x_col, ax=axes[i])
fig.delaxes(axes[-1])
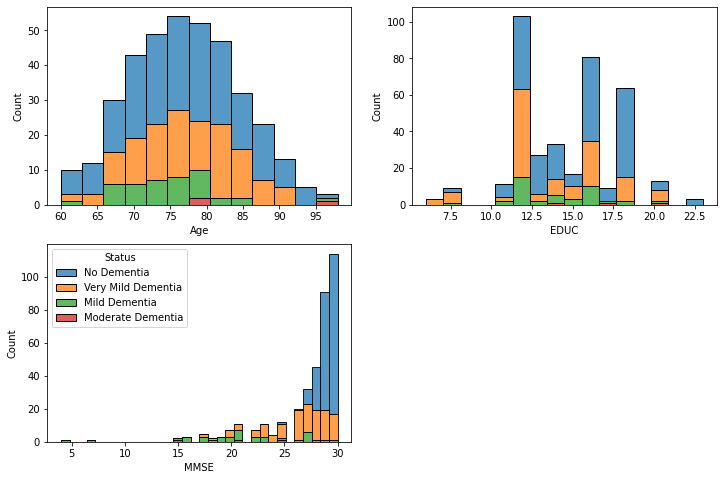
# Histograms stratified by dementia status
fig, axes = plt.subplots(2, 2, figsize=(12,8))
# Make it easier to loop over
axes = axes.flatten()
# Loop through the columns we want to plot a histogram for
for i, x_col in enumerate(["Age", "EDUC", "MMSE"]):
sns.histplot(data=df, x=x_col, hue="Status", multiple="stack", ax=axes[i])
# Manually tidy up the legends
axes[0].get_legend().remove()
axes[1].get_legend().remove()
# Delete the extra axis (as we only plot 3 columns)
fig.delaxes(axes[-1])
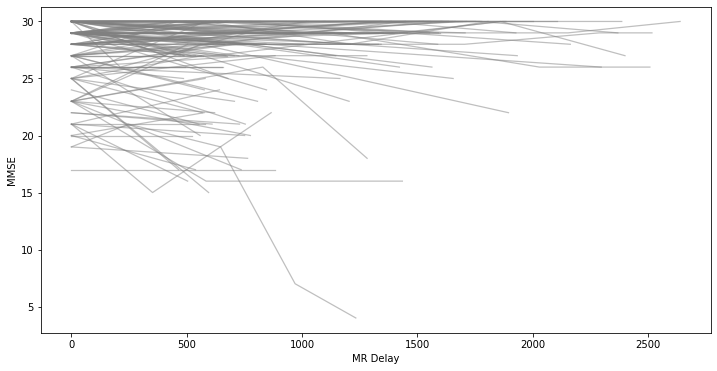
MMSE trajectories over time¶
We can visualize the MMSE trajectories of each subject over time to get an overall view of the cohort.
fig, ax = plt.subplots(figsize=(12,6))
for _, indiv_data in df.groupby("Subject ID"):
ax = sns.lineplot(
data=indiv_data,
x="MR Delay",
y="MMSE",
ax=ax,
color="grey",
alpha=0.5,
linewidth=1.25,
linestyle="solid"
)
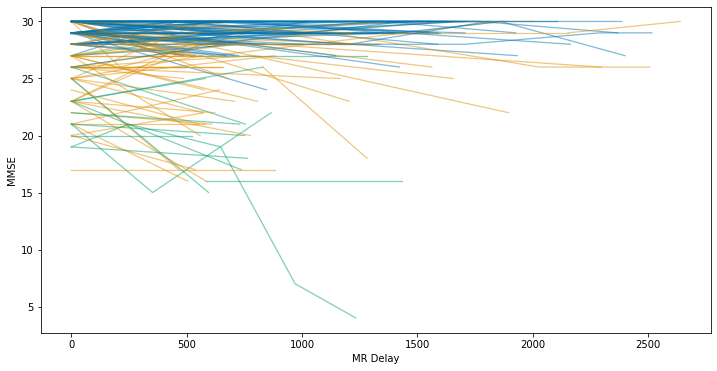
We can make this graph a little more interesting by adding colour to each line, corresponding to the dementia status:
fig, ax = plt.subplots(figsize=(12,6))
# Loop over the individual subjects
for group, indiv_data in df.groupby(["Subject ID"]):
# Extract xy data
xy = indiv_data[["MR Delay", "MMSE"]].values
# Plot normally for patients that do not change status
if len(indiv_data["Status"].unique()) == 1:
ax.plot(
xy[:, 0],
xy[:, 1],
color=status_colours[indiv_data["Status"].unique()[0]],
alpha=0.5,
linewidth=1.25
)
else:
# Duplicate points to construct line segments of different colours
for i, (start, stop) in enumerate(zip(xy[:-1], xy[1:])):
x, y = zip(start, stop)
ax.plot(
x, y,
color=status_colours[indiv_data["Status"].iloc[i]],
alpha=0.5,
linewidth=1.25
)
# Add the labels
ax.set_xlabel("MR Delay")
ax.set_ylabel("MMSE")
Text(0, 0.5, 'MMSE')
Regression Analysis¶
Simple Linear Regression¶
# Select the columns to iterate over
x_cols = ["Age", "Sex", "EDUC", "CDR"]
# Choose nicer names to display for each column
x_labels = ["Age", "Sex", "Yrs of Education", "CDR"]
fig, axes = plt.subplots(2,2, figsize=(16,10))
# Select the dependent variable
y = df_baseline["MMSE"].values
# Loop over the columns to fit each regression
for x_col, label, ax in zip(x_cols, x_labels, axes.flatten()):
# Select x data
x = df_baseline[x_col].values
# Create the model
model = sm.OLS(
y, sm.add_constant(x)
)
# Fit the model
res = model.fit()
# Add a plot with regression line
ax.plot(x, res.predict())
# and the raw data
ax.scatter(x, y, c=[status_colours[i] for i in df_baseline["Status"]])
ax.set_title(label)
ax.set_ylabel("MMSE")
# Print the R^2
print(f"{label} vs MMSE: R-squared = {res.rsquared:.3f}")
Age vs MMSE: R-squared = 0.001
Sex vs MMSE: R-squared = 0.048
Yrs of Education vs MMSE: R-squared = 0.047
CDR vs MMSE: R-squared = 0.479
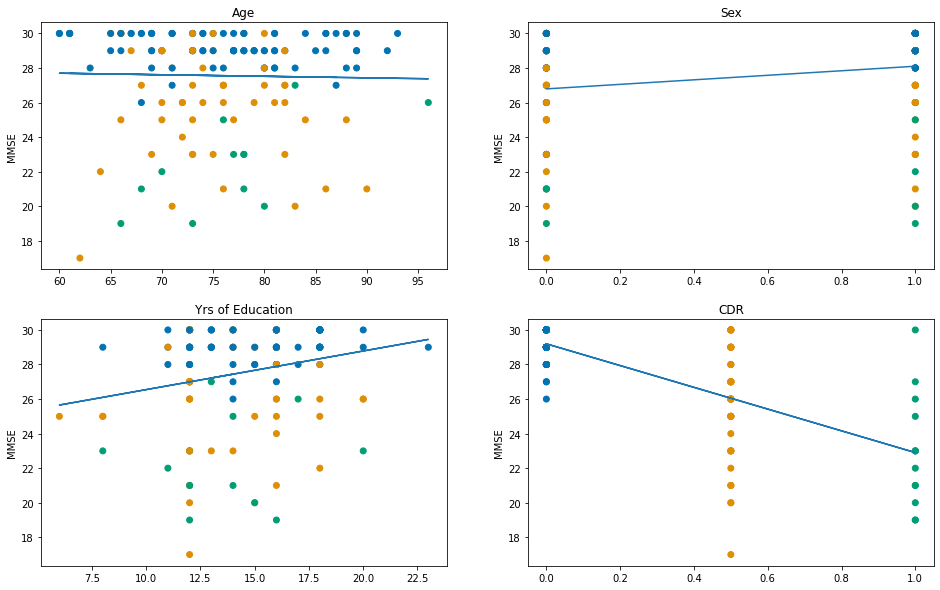
Then we can do the same again, but this time stratify by the dementia status (not including CDR this time):
x_cols = ["Age", "Sex", "EDUC"]
x_labels = ["Age", "Sex", "Yrs of Education"]
fig, axes = plt.subplots(2,2, figsize=(16,10))
axes = axes.flatten()
for x_col, label, ax in zip(x_cols, x_labels, axes):
for name, group_data in df_baseline.groupby("Status", observed=True):
y = group_data["MMSE"]
x = group_data[x_col]
model = sm.OLS(
y, sm.add_constant(x)
)
res = model.fit()
ax.plot(x, res.predict())
ax.scatter(x, y, color=status_colours[name])
ax.set_title(label)
ax.set_ylabel("MMSE")
# Delete the extra axis (as we only plot 3 columns)
fig.delaxes(axes[-1])
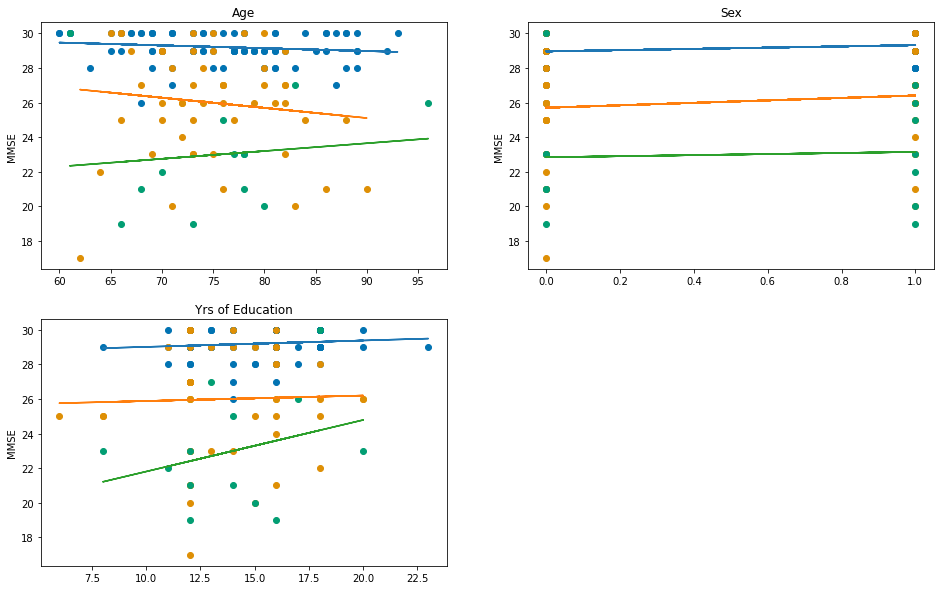
Multiple Linear Regression¶
We can create a regression model using age, sex, education, and CDR as our independent variables together:
# Create string for our independent variables
x = "Age + EDUC + Sex + C(CDR)"
# And select the dependent one
y = "MMSE"
mod = smf.ols(formula = f"{y} ~ {x}", data=df_baseline)
res = mod.fit()
# Print out a nice summary of results
print(res.summary())
OLS Regression Results
==============================================================================
Dep. Variable: MMSE R-squared: 0.493
Model: OLS Adj. R-squared: 0.475
Method: Least Squares F-statistic: 27.95
Date: Thu, 08 Jul 2021 Prob (F-statistic): 1.02e-19
Time: 10:03:10 Log-Likelihood: -324.67
No. Observations: 150 AIC: 661.3
Df Residuals: 144 BIC: 679.4
Df Model: 5
Covariance Type: nonrobust
=================================================================================
coef std err t P>|t| [0.025 0.975]
---------------------------------------------------------------------------------
Intercept 29.1743 2.140 13.632 0.000 24.944 33.404
C(CDR)[T.0.5] -2.9600 0.411 -7.209 0.000 -3.772 -2.148
C(CDR)[T.1.0] -6.0383 0.649 -9.305 0.000 -7.321 -4.756
Age -0.0194 0.024 -0.825 0.411 -0.066 0.027
EDUC 0.0734 0.065 1.137 0.257 -0.054 0.201
Sex 0.5643 0.374 1.511 0.133 -0.174 1.303
==============================================================================
Omnibus: 23.128 Durbin-Watson: 1.929
Prob(Omnibus): 0.000 Jarque-Bera (JB): 53.918
Skew: -0.623 Prob(JB): 1.96e-12
Kurtosis: 5.660 Cond. No. 943.
==============================================================================
Warnings:
[1] Standard Errors assume that the covariance matrix of the errors is correctly specified.
# Call statsmodel function for partial regression plot
fig = sm.graphics.plot_partregress_grid(res)
# Making the output nicer
fig.set_figheight(10)
fig.set_figwidth(12)
fig.tight_layout(pad=3)
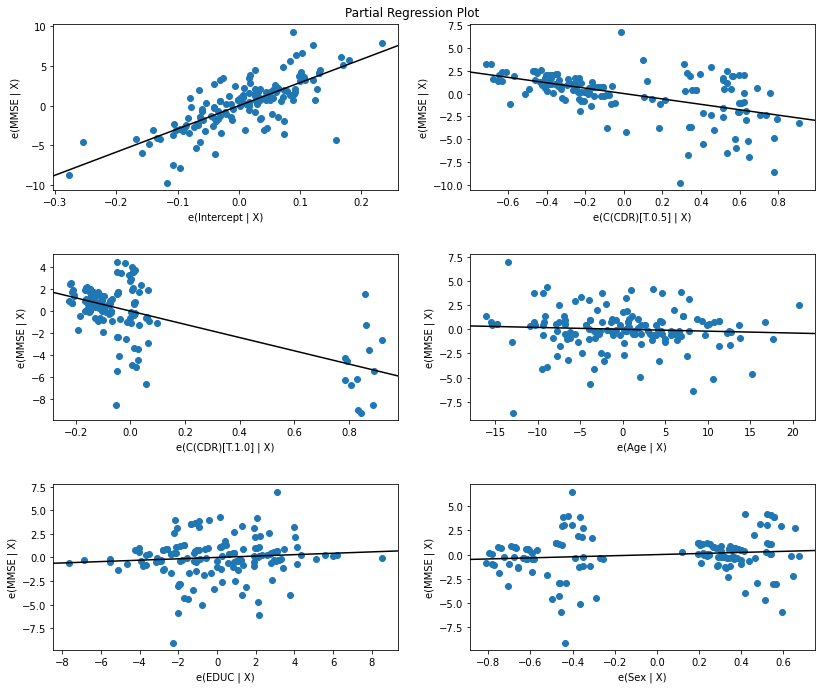
Results Table¶
Either using some of the analysis above, or through additional analysis of your own, create a summary results table below. What it includes is up to you, the overall aim is to get familiar with creating tables in markdown.
Export to PDF¶
Now we can export this report to PDF, using the steps below.
Note: You may need to change the paths and filenames depending your Google Drive structure, and what you have named this notebook.
# Install what we need to create the PDF
%%capture
!apt update
!apt install texlive-xetex texlive-fonts-recommended texlive-generic-recommended
# Mount Google Drive
from google.colab import drive
drive.mount('/content/gdrive')
Drive already mounted at /content/gdrive; to attempt to forcibly remount, call drive.mount("/content/gdrive", force_remount=True).
# Create a DEMON folder and move this notebook into it
%%bash
mkdir -p gdrive/MyDrive/DEMON/
if [ -f gdrive/MyDrive/Reproducible_Reporting.ipynb ]; then
mv gdrive/MyDrive/Reproducible_Reporting.ipynb gdrive/MyDrive/DEMON/Reproducible_Reporting.ipynb
fi
ls gdrive/MyDrive/DEMON/
Reproducible_Reporting.ipynb
# Change to the drive we created above
%cd gdrive/MyDrive/DEMON/
/content/gdrive/MyDrive/DEMON
!jupyter nbconvert Reproducible_Reporting.ipynb --to pdf --output ./Reproducible_Reporting.pdf
[NbConvertApp] Converting notebook Reproducible_Reporting.ipynb to pdf
[NbConvertApp] Writing 49953 bytes to ./notebook.tex
[NbConvertApp] Building PDF
[NbConvertApp] Running xelatex 3 times: [u'xelatex', u'./notebook.tex', '-quiet']
[NbConvertApp] Running bibtex 1 time: [u'bibtex', u'./notebook']
[NbConvertApp] WARNING | bibtex had problems, most likely because there were no citations
[NbConvertApp] PDF successfully created
[NbConvertApp] Writing 59264 bytes to ./Reproducible_Reporting.pdf